Mass Spectrometry Immunoassay (MSIA) Facility
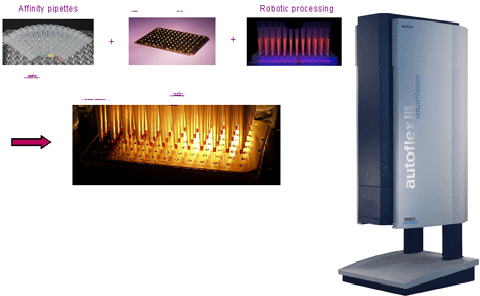
Key System Highlights:
- Automated (robotic) protein biomarker validation
- Characterization of bio therapeutic drugs in clinical trials
- Coupled antibody affinity capture to mass spectrometry for quantitative and qualitative multiplexed immunoassays
- Distinguishes changes in proteins arising from amino acid substitutions, RNA splice variants, posttranslational modifications, proteolytic cleavage and bound molecules
- 500 samples/day throughput
- Initial menu of over 75 assays currently available
- Customized assays for other proteins defined by client
Contact Information:
Seema Husain, Ph.D.973-972-3170
husainse@njms.rutgers.edu
Background:
One of the major limitations of protein studies is the scarcity of high throughput technologies to simultaneous measure and readily recognize and characterize micro heterogeneity that is encountered in human populations that can lead to disease. Mass spectrometry and database search engines are commonly used to identify proteolytic fragments that are purposely created as surrogates of the proteins in a sample. But in doing so, full-length protein information is lost during proteolytic digestion thus eliminating the detection of unique protein characteristics within an individual stemming from genetic, transcriptional and posttranslational differences. Likewise, most conventional immunoassay approaches (e.g., ELISA and RIA) cannot differentiate structural changes, and are thus unable to recognize diversity and modifications at the protein level. The problem is further exacerbated when considering the dynamics of disease-states, where specific proteins are modulated, both quantitatively and qualitatively, within a disease population.
In 1995, Intrinsic Bioprobes Inc. (IBI) introduced the Mass Spectrometric Immunoassay (MSIA) as a technique for detecting and identifying peptides and proteins present in biological fluids [1]. This proprietary approach [2] is based on the isolation of analytes from biological milieu using immobilized antibodies, followed by release of the analytes and analysis using MALDI-TOF-MS. The original papers and patents describe methods for qualitative determination and identification of analytes, quantitative methodologies and multiplexed MSIA where multiple antibodies are used to target multiple analytes simultaneously. To simplify the approach IBI introduced the use of solid supports built into pipette tips for protein isolation prior to MALDI-TOF-MS (termed MSIA-Tips) [3]. These devices can address large volumes of bio fluid and are used with 96-well (parallel) robotic workstations that can reach throughputs of ~500 assays/day [4].
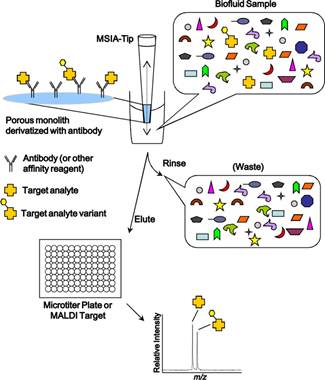
Hussein Yassine, et al. Proteomics Clin Appl. ;7(0):528-540
General scheme of mass spectrometric immunoassay.
This system couples automated antibody affinity capture to mass spectrometry, allowing determination in one platform of both sensitive quantification with fmol sensitivity as well as qualitative changes in protein structure arising from amino acid substitutions, RNA splicing, posttranslational modifications, proteolytic processing and binding partners. This technology has been well established in industry for over 10 years, used for biomarker validation and characterization as well as characterization of biotherapeutic drugs after they are given to patients in clinical trials.
The MSIA platform is particularly well suited for screening plasma and urinary proteins for micro heterogeneity, and as such is a technique that fills the aforementioned "analytical void" existing between global proteomics approaches and conventional immunoassays. IBI has applied this technology to develop over 75 proprietary assays, used to target specific proteins for rigorous structural characterization and quantification.
An example of the resolution of this technology is exemplified above, in a typical mass spectrum resulting from the cystatin C mass spectrometric immunoassay of human plasma. Cystatin C, a cysteine proteinase inhibitor and a potential marker for several pathological processes. Beta lactoglobulin, an internal reference standard was incorporated into the assay, servingas a normalization point for cystatin C quantification. Signals for native CysC (MW = 13,343) along with several cystatin C variants: CysC containing hydroxyproline at position 4 (CysC 3Pro-OH; MW = 13,359),
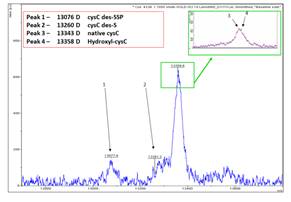
Mass spectrum of modifications detected in Cystatin C.
CysC missing its N-terminal Serine (CysC des-S, MW = 13,260), and CysC missing its three N-terminal residues (CysC des-SSP, MW = 13,076) are detected. The assay determines the concentration of cystatin C variants across a cohort of samples, demonstrating the ability to fully quantify individual forms of post-translationally modified proteins [5].
1. Nelson RW, Krone JR, Bieber AL, Williams P. 1995. Anal. Chem. 67: 1153-8.
2. The Mass Spectrometric Immunoassay (MSIA) process is patented under U.S. Patent #6,974,704, which is assigned with exclusive rights to Intrinsic Bioprobes, Inc. Tempe, AZ.
3. These devices and their use in HT-MSIA are protected under U.S. Patent #'s, 6,004,770; 6,093,541; 6,316,266; 6,498,039; 6,783,672; 7,083,723; 7,083,724; 7,087,163; 7,087,164; 7,087,165, all of which are assigned with exclusive rights to Intrinsic Bioprobes, Inc. Tempe, AZ.
4. Kiernan, U.A., Tubbs, K.A., Gruber, K., Nedelkov, D., Niederkofler, E.E., Williams, P., Nelson, R.W., "High Throughput Protein Characterization using Mass Spectrometric Immunoassay", Anal. Biochem. 2002; 301(1):49-56>.
5.Trenchevska O1, Kamcheva E, Nedelkov D., Mass spectrometric immunoassay for quantitative determination of protein biomarker isoforms, J Proteome Res. 2010; 9(11): 5969–5973.

 Institute of Genomic Medicine
Institute of Genomic Medicine